|
CCMB |
 |
RCC |
 |
|---|
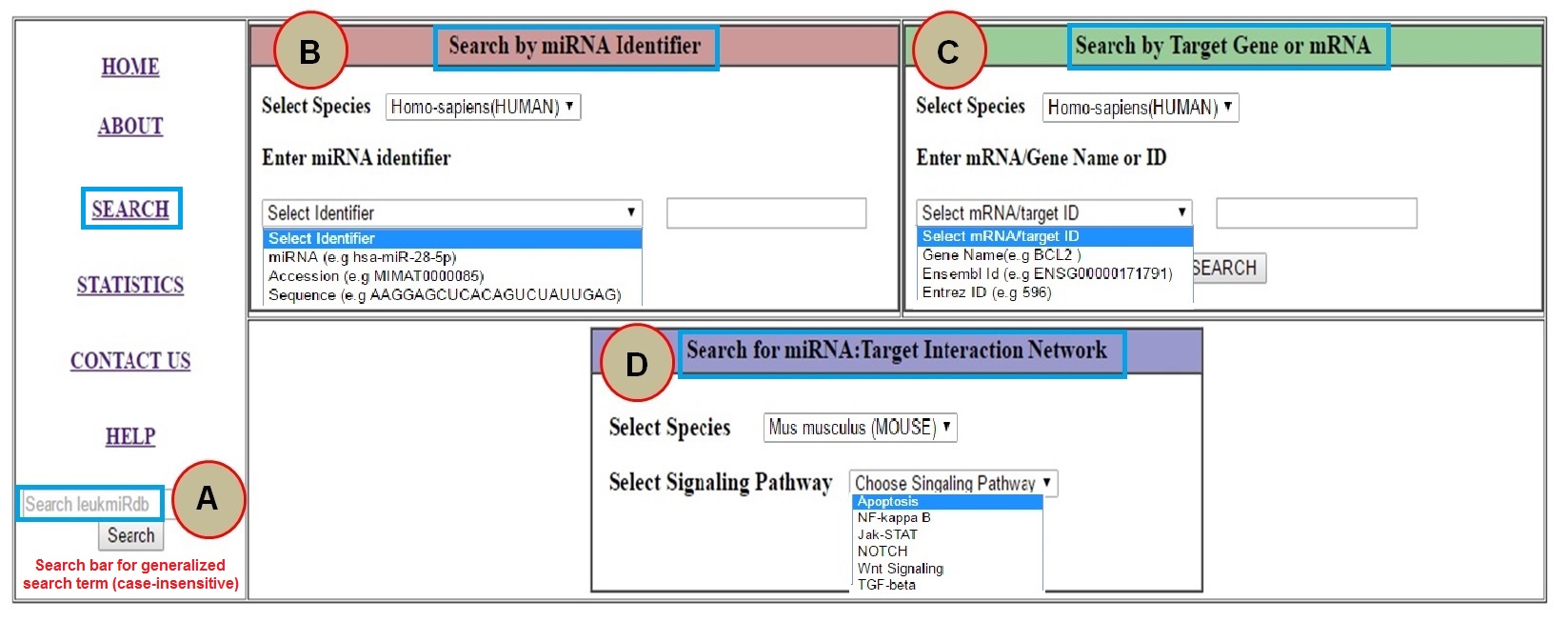
How to explore LeukmiR database? The HOME page and the SEARCH button provides interactive user friendly database searching environment to users in multiple ways.
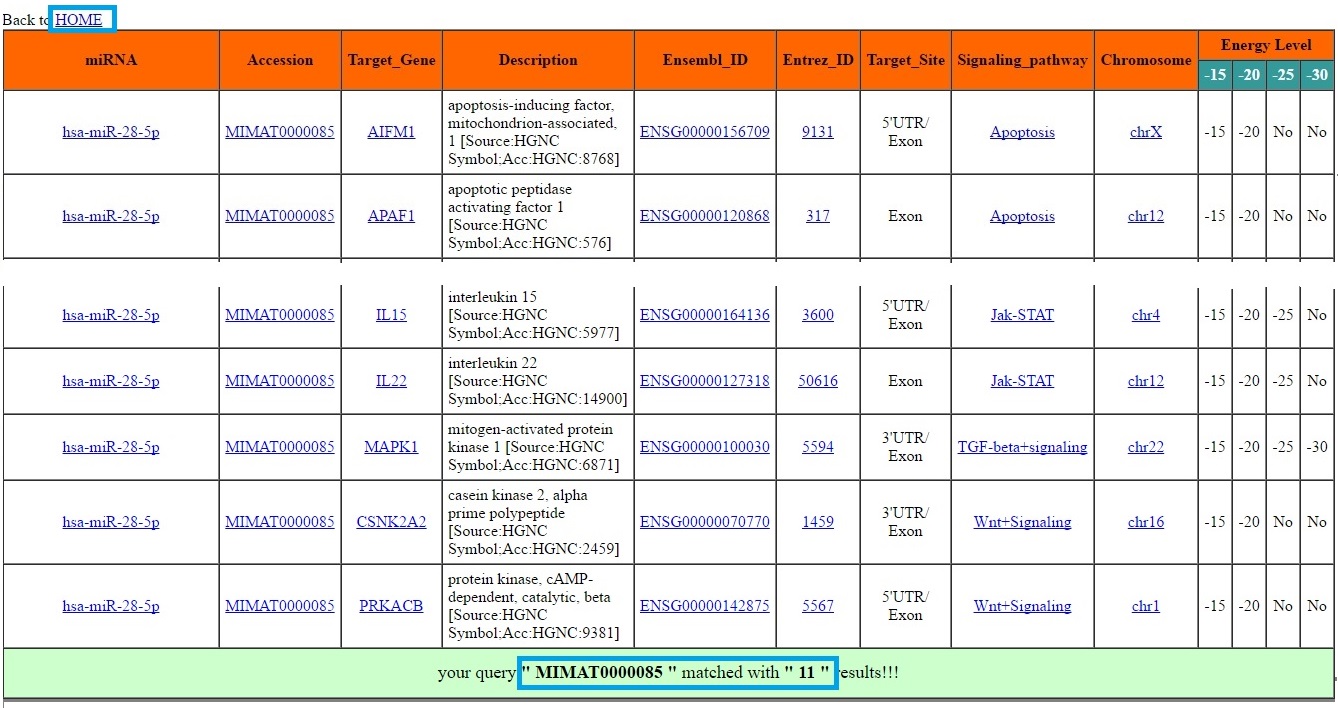
The Searchbar provided on HOME page can be use for generalise search purpose. User can do any search query related to miRNAs and their targets which is case-insensitive such as Species name (homo-sapiens or mus-musculus), miRNA name, miRNA sequence, miRNA accession, target gene, Entrez-ID etc. The output will be shown in new windows stating the total number of results matched with your query. Result Window: 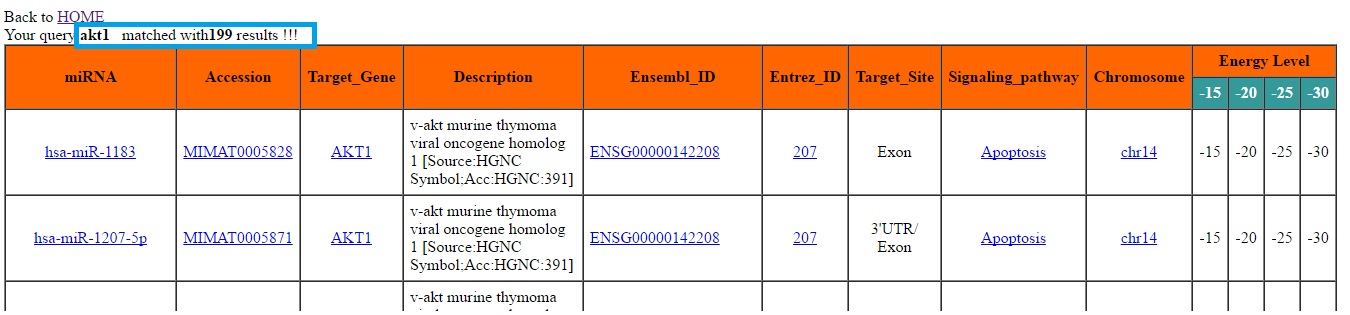
The SEARCH button provides three quite different search option. User can use either miRNA identifier based search option or mRNA (target gene) based search option for any one of the species (Human/Mouse) at a time. The miRNA based search can be done using different miRNA identifier such miRNA name (eg. hsa-miR-28-5p), miRNA miRBase accession ID (eg. MIMAT0000085) or using mature miRNA sequence (e.g. AAGGAGCUCACAGUCUAUUGAG). In similar way the user can opt mRNA or target gene based search option to see what are the miRNAs targeting the particular gene in any one of the specified genome (Human/Mouse). User can search using different gene identifiers such as Gene name (eg. BCL2), entrez ID (eg. 596) or Ensembl ID (eg. ENSG00000171791) Result Window:
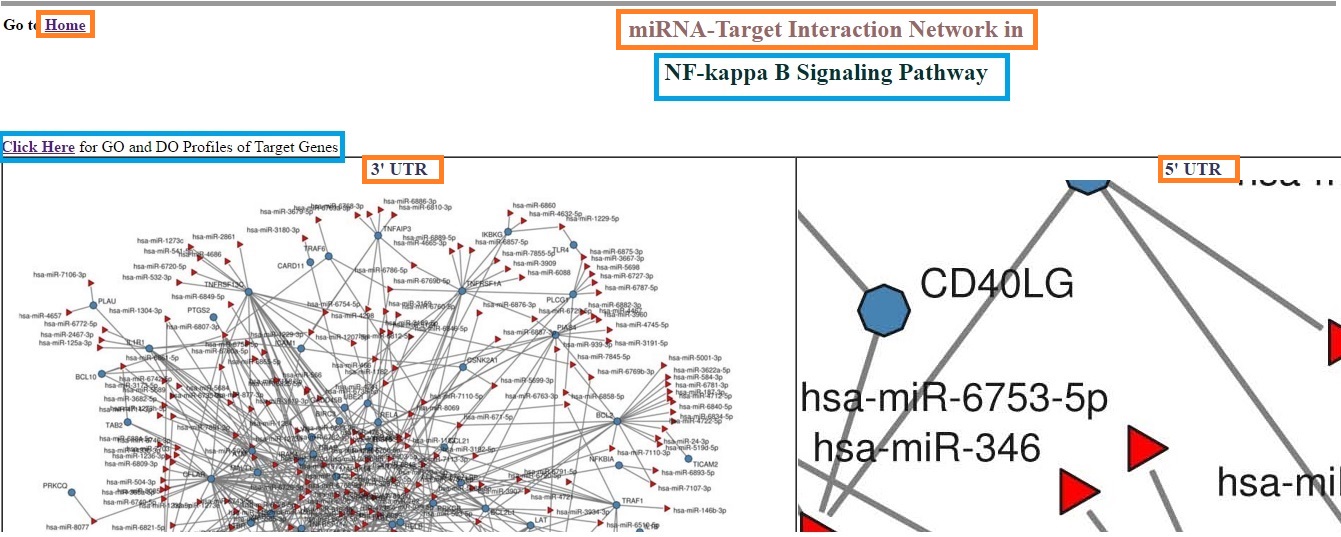
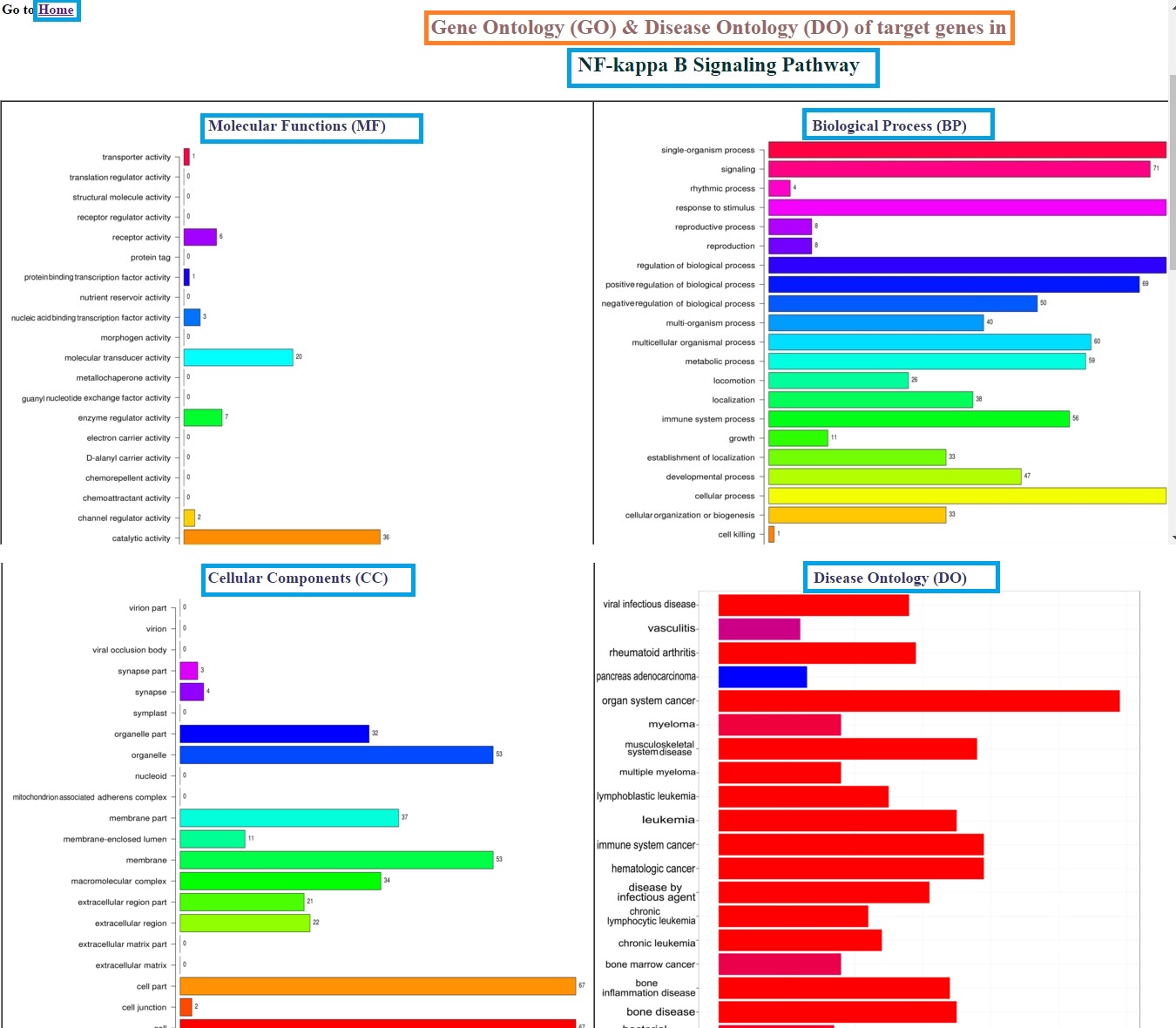
User can also explore the miRNA:mRNA interaction network in different signaling pathways for both Human and Mouse Genome. The output window will show miRNA:mRNA interaction network at three different genomic target site i.e 3'UTR, 5'UTR of their target genes in a signaling pathway. User can zoom the network by scrolling the cursor over network. The pathway interaction page also provides Gene Ontology (GO) and Disease Ontology (DO) profiling of target genes in particular signaling pathway. Result window:
GO and DO profiles Result Window:
|
|---|